Structure/function of cytochrome P450 enzymes; drug metabolism; drug design
While most enzymes are very specific for a single substrate, a few are much more versatile. The main interest of the Scott laboratory involves the ability of cytochrome P450 (CYP) enzymes to both bind and metabolize a number of chemicals that are very different in size, shape, and stereochemistry. Within an organism, the presence of a set of these multi-taking enzymes is largely responsible for the ability to eliminate foreign chemicals including drugs, environmental substances, and carcinogens.
Major questions include:
- How does the three-dimensional structure of a cytochrome P450 recognize, bind, and metabolize multiple chemically diverse substrates?
- By what mechanism can two different P450 enzymes act on a common substrate but differentiate between other chemicals that only one of them can metabolize?
- How do selective inhibitors interact with the protein structure in these versatile enzymes?
- How can we use this knowledge to advance human health by preventing disease, understanding how drugs are manipulated in the body, and to treat diseases?
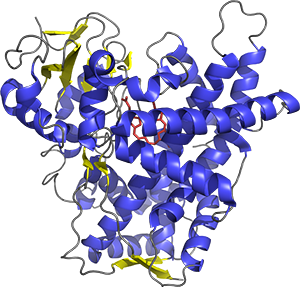
To investigate these and related questions, the laboratory uses methods in molecular biology, biochemistry, X-ray crystallography, and NMR. Recently major projects in the lab have involved the human P450 enzymes 2A6, 2A13, 2E1, and 1A1, which constitute a model system for examining enzymes with both overlapping and distinct substrates. The general hypothesis is that comparison of molecular structure and biochemistry across different P450 enzymes will assist in the identification of physical and chemical protein features responsible for differences in their function. Theses characteristics can then be used to understand and predict substrate specificity and to design selective inhibitors for clinical use.
Applications to prevention of lung cancer: CYP2A13
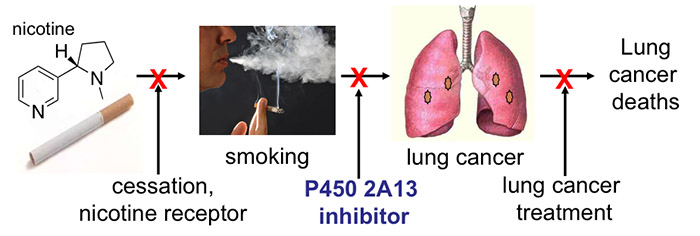
One application of the basic biochemistry above is our effort to design a selective inhibitor of cytochrome P450 2A13 for the prevention of lung cancer in smokers. CYP2A13 acts on a ring-opened form of nicotine to clear it from the body. Unfortunately, in the process two carcinogens are created by CYP2A13. These carcinogens interact with DNA to cause lung cancer. Since smokers without a functional copy of the CYP2A13 protein are physiologically normal but less likely to develop lung cancer, we reasoned that inhibition of CYP2A13 activity could be used to safely reduce lung cancer risk in those who cannot or will not give up exposure to nicotine. Working with the KU High Thoughput Screening Laboratory and the KU Specialized Chemistry Center we have developed compounds that selective inhibit CYP2A13, but not the closely related human liver CYP2A6 enzyme. These compounds are currently being tested to determine if they will make good pharmaceutical agents.
Applications to prostate and breast cancer: CYP17A1
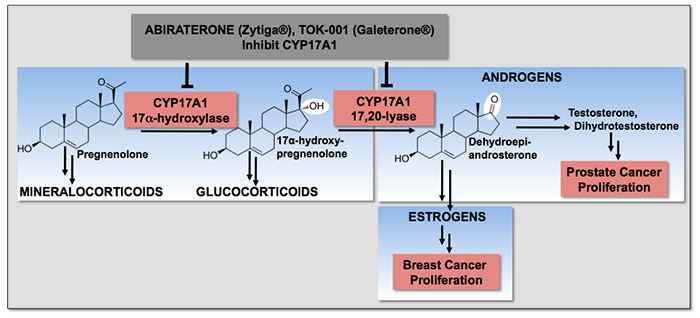
Cytochrome P450 17A1 (CYP17A1) performs sequential hydroxylase and lyase reactions essential for the production of androgen and estrogen sex steroids. Prostate cancer and breast cancer proliferate in response to androgens and estrogens, respectively. Thus inhibition of CYP17A1 is a new approach to treat prostate and breast cancer, but no structures of CYP17A1 were previously available to assist in drug design. We determined structures of CYP17A1, first as complexes with drugs approved by the FDA for metastatic prostate cancer (abiraterone or Zytiga(R)) or currently in clinical trials (TOK-001 or Galeterone(R)). These structures:
- demonstrated that both drugs bind almost perpendicular to the heme prosthetic group, rather than the parallel orientation predicted
- revealed important opportunities to improve drug design, which are the subject of ongoing research
- demonstrated similarities to steroid receptors likely to form the basis for the dual mechanism of action for TOK-001 at the androgen receptor
- elucidate the structural roles of many mutations found in patients with diseases called 17-hydroxylase deficiencies